Paper in Nature: De novo protein design
29 juli 2019 - Harald Feldmann
29 juli 2019 - Harald Feldmann
Advances in de novo proteïn design over the past 10 years
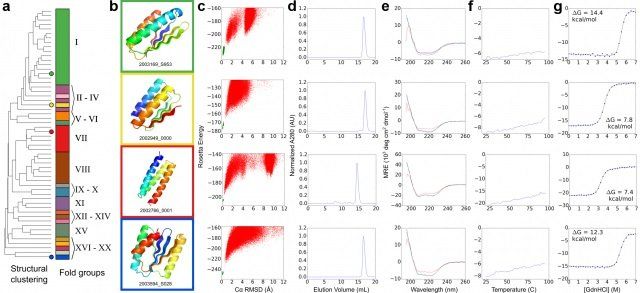
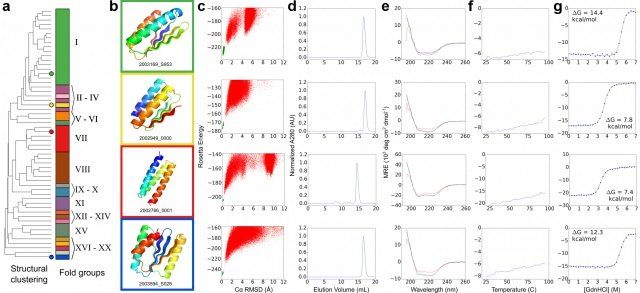
Over the past 10 years proteïns have been designed in the Fold.it project. In this process a string of beads, as it were, is designed using beads in such a way that the string takes on a specific shape. It is a very elaborate process in which promising designs are first analyzed statistically. This is done by processing the model by a network of more than 40.000 computers worldwide, the so-called 'Rosetta at home' network. The result of that first analysis are the red graphs in the image above.
If the red graph takes on a characteristic plume shape, things get serious. Because the chemical sequence of the design, the sequence of the beads, is known, fragments of DNA can be ordered so that, when put together into a bacterium, they will spell the 'blueprint' of the designed model. Such a piece of DNA can be compared to a letter in a letterbox which, when placed in the right order, spells the desired word. In this case the DNA pieces were inserted into the Escherichia coli (E.coli) bacterium. This modified bacterium will then start to produce the chemical substance that was designed on the computer ('spelling' the desired 'word').
When there is enough of the material collected (enough copies of the 'word' created), the material is crystallized. Think of a salty solution that is being evaporated, the salt remains. The crystal now is bombarded with x-rays to determine the 3d structure. In a sense an x-ray is taken of the crystal. In the paper mentioned here, 146 models were tested this way. 56 of those not only turned out to be exactly as designed but also remained stable when put in liquid.
Harald Feldmann has worked on 5 of these successful, stable, designs.
Read the entire article for free online: https://rdcu.be/bFE7R
Read the entire article on the Nature website: https://www.nature.com/articles/s41586-019-1274-4
If the red graph takes on a characteristic plume shape, things get serious. Because the chemical sequence of the design, the sequence of the beads, is known, fragments of DNA can be ordered so that, when put together into a bacterium, they will spell the 'blueprint' of the designed model. Such a piece of DNA can be compared to a letter in a letterbox which, when placed in the right order, spells the desired word. In this case the DNA pieces were inserted into the Escherichia coli (E.coli) bacterium. This modified bacterium will then start to produce the chemical substance that was designed on the computer ('spelling' the desired 'word').
When there is enough of the material collected (enough copies of the 'word' created), the material is crystallized. Think of a salty solution that is being evaporated, the salt remains. The crystal now is bombarded with x-rays to determine the 3d structure. In a sense an x-ray is taken of the crystal. In the paper mentioned here, 146 models were tested this way. 56 of those not only turned out to be exactly as designed but also remained stable when put in liquid.
Harald Feldmann has worked on 5 of these successful, stable, designs.
Read the entire article for free online: https://rdcu.be/bFE7R
Read the entire article on the Nature website: https://www.nature.com/articles/s41586-019-1274-4

Buitengewoon vereerd te hebben bijgedragen aan het succes van Fold.it en de makers van AlphaFold te hebben geinspireerd tot hun baanbrekende werk op het gebied van Computational Protein Design en Protein Structure Prediction ! Geweldig nieuws voor iedereen en een stimulans om bij te dragen aan nieuwe onderzoeken en technieken door de Truus Schut Stichting. Felicitaties aan David Baker, Demis Hassabis en John M. Jumper !

In een lezing voor de Universiteit van Oxford geeft Dr. Demis Hassabis, CEO van Google Deepmind en bedenker van het programma AlphaFold, credit aan ons werk op het gebied van Computational Protein Design en vermeldt daarbij expliciet hoe de papers in Nature en Nature Structural (and Molecular) Biology waar Harald Feldmann co-auteur van is hem hebben geinspireerd om AlphaFold te maken. Dit is een groot compliment voor al het harde werk dat de afgelopen 15 jaar is verricht en doet eer aan de bijdrage die Harald Feldmann daarmee gedaan heeft aan het werk van Dr. David Baker, bedenker van Fold.it en director van het Institute for Protein Design aan de Universiteit van Washington.

Vereerd door Fold.it's keuze ' Ontwerp van de maand'. Een door Harald Feldmann ontworpen NTF2 ligand binder. Deze structuur is deels ontworpen met behulp van A.I. en vervolgens verder ontwikkeld met zelf ontworpen software op het cluster van de Stichting. De complexiteit van vorm en samenstelling zijn reden om deze volledig nieuwe, niet in de natuur voorkomende, structuur 'ontwerp van de maand' te maken. De structuur zou gebruikt kunnen worden om bepaalde medicijnen, z.g. 'liganden', in te 'verpakken' voor toediening in het lichaam.
Proteïnes willen zich graag in een toestand van zo laag mogelijke energie vouwen. Bij proteïne ontwerp wordt via software gezocht naar een toestand van zo laag mogelijke energie, maar door beperkingen in de algoritmes kunnen soms lokale minima worden gekozen in plaats van het absolute minimum. Stel je een meertje in de bergen voor waarvan je denkt dat het een meer op zeeniveau is. Door een landkaart te maken van de hele omgeving kom je erachter welke meertjes er nog meer zijn en waar uiteindelijk de zee is. Een Machine Learning methode die dit kan doen voor proteïne ontwerp (het in kaart brengen en optimaliseren van het energie landschap) is thema van de publicatie 'Protein sequence design by conformational landscape optimization' in de 'Proceedings of the National Academy of Sciences', PNAS: https://www.pnas.org/doi/10.1073/pnas.2017228118 waar Harald Feldmann aan heeft meegewerkt.
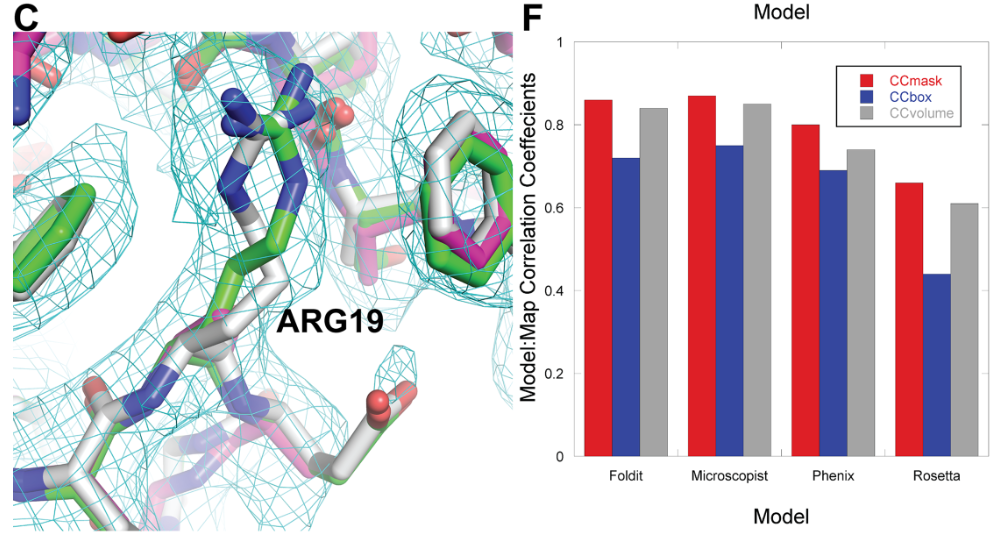
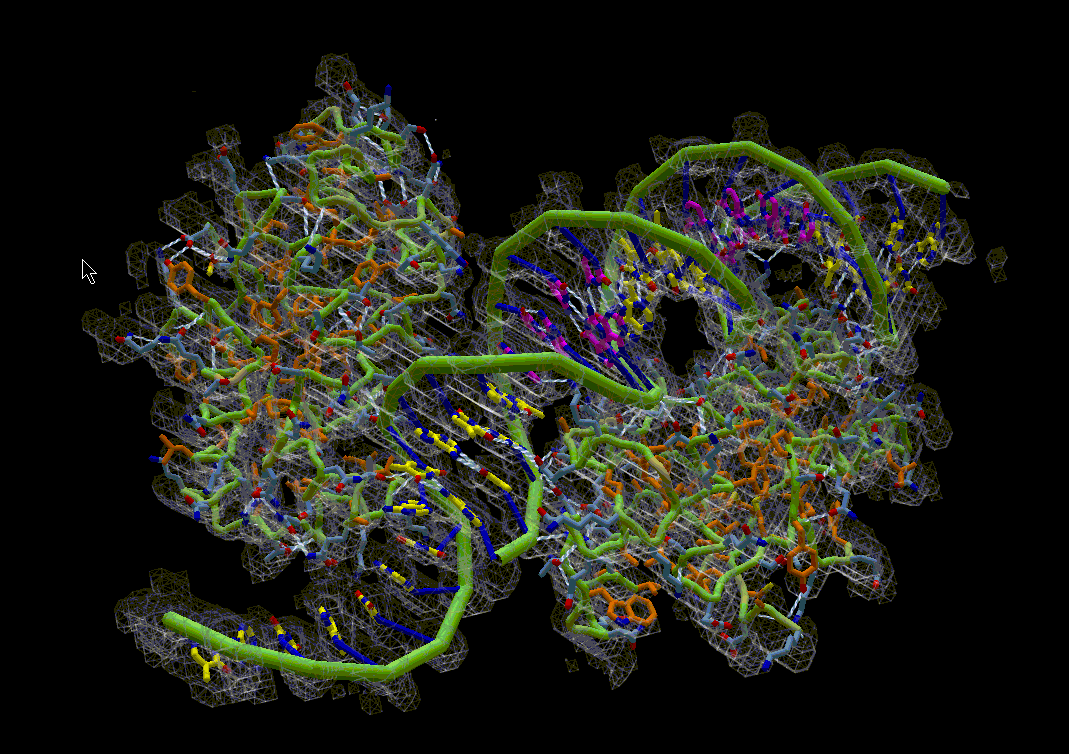
Microscopen kunnen kleine objecten bekijken met behulp van licht. Electronen Microscopen kunnen héle kleine structuren bekijken met behulp van een electronenbundel. Proteïne structuren zijn hele kleine structuren en door ze heel sterk af te koelen kan men een globale structuur bepalen met een Electronen Microscoop. Dat afkoelen gaat tot 4 graden boven het absolute nulpunt, of ongeveer -269 graden Celsius. Dat noemt men 'Cryogeen'. Hier komt "Cryo-" vandaan uit Cryo-EM, het cryogeen afkoelen voor gebruik in een electronen microscoop. Om de structuur die men hiermee kan vaststellen te verfijnen zijn algoritmes bedacht die door computers kunnen worden gebruikt. De beste algoritmes zijn echter nog niet zo goed als een expert in het bepalen van de beste match tussen een 'wolk' uit de Cryo-EM meting en een echte structuur. In een experiment werd een proteïne structuur globaal vastgesteld in een Cryo-EM meting. Die werd vervolgens aangeboden aan microscoop specialisten, diverse algoritmes en de Foldit groep. In deze paper, waarvan Harald Feldmann als lid van de Foldit groep co-auteur is, wordt uitgelegd hoe de verschillende groepen de klus hebben geklaard. Het blijkt dat een aantal personen uit de Foldit groep de beste structuren heeft gemaakt. Een hele prestatie ! Lees de paper online: https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000472